Gallery
Gallery
Tumor Classification via High Resolution Digital Image Tiling
Media Details
Created 07/20/2004
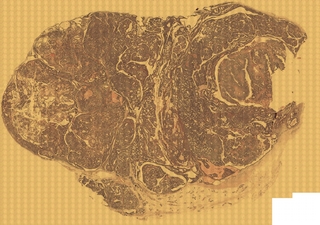
Tumor slices were obtained from rats sacrificed using 2 mL sodium pentobarbital (Nembutal) IP and transcardially perfused with 0.1 M phosphate buffer (PB), pH = 3.5. The tumors were cross sectioned based on scout images, which maintained the orientation planes of the tumor in situ. The tissue slices were sectioned at a thickness of 3[mu]m, and stained with hemotoxylin and eosin (HE) for routine histopathology. Dr. Keith Bailey, College of Veterinary Medicine, Department of Pathobiology, UIUC performed the sectioning, staining, and tumor classification. High resolution digital images of the tumor slices were created with the Microscopy Suite's fluorescence microscope using a 20x objective and the MCID image tiling software. This created a series of image tiles which were then aligned and stitched in the VMIL into a 5.74GB image (54,055x38,037 pixels) using custom software written by ITG. A much-reduced copy of that image is reproduced here. The histiopathologically determined cellularity and capillary density will be compared with dynamic contrast enhanced (DCE) MRI-based maps of two-compartment model parameters, namely tumor extravascular extracellular space (EES) volume fraction (vc), tumor plasma volume fraction (vp), and volume normalized contrast agent transfer rate between tumor plasma and EES (Kpt/Vt). This comparison will be performed in the VMIL using various software packages for image analysis.
Credits
- Dr. Keith Bailey , Nuclear Engineering
- Michael Aref , Nuclear Engineering
- Amir Chaudhari , Nuclear Engineering